Abstract
By Brigette Corder, Sterling Ericsson, and Taylor Uhlir
The CRISPR-Cas systems are a new and exciting tool for research and scientific discovery. Here we discuss and compare the various CRISPR-Cas systems and report current uses for these systems by concentrating a principal spotlight on CRISPR-Cas 10. From CRISPR’s rudimentary beginnings in the form of the Cascade system to the expanded motley of specialized, but diverse collections of Cas proteins, the CRISPR-Cas systems have been found to incorporate complexes with many capabilities. This includes differing forms of genetic repair and even targeted subunits directed at RNA cleavage rather than the common DNA default.
The structure of CRISPR-Cas 10 in particular has several key differences from the better known CRISPR-Cas 9 system. These differences relate directly to the function of CRISPR-Cas 10. Specifically, it does not require PAM sequences and shows promise for identifying sequences even with point mutations. In light of this, the CRISPR-Cas 10 system shows special potential for bioengineering and gene editing.
The Structure and Potential of the CRISPR Cas 10 System
The diversity of CRISPR-Cas systems spans a spectrum of targeted threats and cross-purposes unique to many bacterial families. Thanks to the individualized evolutionary pressures on each, disparate CRISPR proteins have changed over time to create an entire menagerie of different answers to the question of viral invasion defense. While some species retain what are nominally considered more primitive forms of CRISPR defenses, others have randomly, but mechanistically evolved to incorporate a more advanced complex.
The following will serve as a gateway to the structure of Cas proteins, their purposes, and how the variety of CRISPR systems are organized by contemporary science, even as ongoing rearrangements and additions are made by the month. As a bacterial barricade, CRISPR also delivers as an able gene editing tool available for humans to handle in medicine, agriculture, and beyond. Having a thorough comprehension of its makeup serves to enlighten the new possibilities that CRISPR opens to future research and development. Hereafter, we shall focus our discussion on the potential of the CRISPR-Cas 10 system.
An Overview of the CRISPR System and CRISPR Protein Complex
To start, the Cas proteins in a CRISPR system are classified into groups referred to as modules based on their activity. The first module is activated when the bacteria comes under threat by a viral target, mobilizing proteins for spacer incorporation. Referred to as adaptation, this module requires destruction of the foreign viral genome and theft of particular sequences from the cleaved remains. These desired sequences are considered protospacers and their insertion into the bacterial genome is mediated by precise proteins (Makarova et al., 2015).
The Cas proteins involved in obtaining spacers are largely conserved across all the divisions. Cas1 and Cas2 are the primary effectors at this stage, with the former acting as an integrase for the actual process of placing the protospacers into the looped spacer sites. The function of Cas2, a homologue for inteferase toxins, is less understood, but what is known is that it complexes with Cas1 and is critical for the catalytic activity needed to insert the protospacers. They can also be joined, in Types I, II, and V, by Cas4, a restriction endonuclease that helps with cleavage of the genome for spacer insertion (Makarova et al., 2015).
The expression and interference modules have a greater variability of involved proteins than the others and are the main reason for the split between Classes of CRISPR Types. Broadly speaking, the effector complex of CRISPR is a multisubunit system with several proteins working in concert to enact the targeting and cleaving processes (Barrangou, 2015). The Expression module includes the production of pre-crRNA (CRISPR RNA) that are then bound to the effector complex and processed into what is considered a mature crRNA. This is commonly done by Cas6 acting as an RNA endonuclease. The complex has at its center a RNA recognition motif (RRM), a required portion needed to bind to viral genes. RRM is a part of the RAMP protein superfamily and shares this distinction with the other common proteins in the complex, Cas5 and Cas7. These act as general non-catalytic structural skeletons for the complex, with Cas7 later working as an RNA degradation component (Hale et al., 2009).
The final segments of the CRISPR complex are the large and small subunits. Their structure is predominantly conserved across Class 1 systems, though the precise gene encoding them varies. Due to this conserved nature, it is believed that the multisubunit effector complex was developed ancestrally before the split into separate Types within Class 1 (Sashital, Wiedenheft, & Doudna, 2012). Once loaded with mature crRNA, the complex is capable of using its RNA-binding domains to trap and interrogate the sequences of targeted DNA (sometimes RNA), starting the interference module. Guide RNAs work as directional components to lead the complex to viral genomes. Once arrived, the previously copied spacers are used to base bind with the sequence and, if a match is located, cleavage factors are initiated (Jinek et al., 2012).
The final ancillary module acts as extra accessory proteins to the overall complex. They are not a part of the core complex and have a higher rarity of inclusion and use in any particular bacterial system. The only common component is Cas4, which acts dually as a part of spacer insertion and as a regulator, currently believed to be involved in using the CRISPR system for programmed cell death under certain circumstances. Other proteins exist outside of the known framework, but few have been characterized or studied to any meaningful depth (Makarova et al., 2015). For a better understanding and breakdown of the Cas proteins, especially in relation to the Types discussed below, refer to Figure 1.

Figure 1: Functional classification of Cas proteins. Designation of Classes, Types, and inherent Cas proteins involved in each module of activity. The Types of CRISPR systems (at left) are split between two Classes due to the broad multifocus proteins found in Class 2. The function and activity of the Cas proteins in each Type are listed (at top) and split between four modules of influence. (Makarova et al., 2015).
Class 1 (Type I [Cascade], Type III [Csm/Cas10, Cmr], Type IV [Csf1])
The first Class of CRISPR systems is usually considered to be the more ancient lineage and the least understood category outside of the original Cascade system, with less streamlined proteins for viral defense. The basic layout of this original Type I Cascade system includes Cas1 through 8, with the subtype varieties involved in differences in operon placement and gene cluster encoding. Some of these also lack Cas4 or have situations where the Cas3 protein is fused to Cas2 (Wiedenheft et al., 2011; Jore et al., 2011).
The standout feature of the Type III systems is the Cas10 protein. The activity of this within Type III-A, Csm (currently just referred to inherently by itself as Cas10), will be discussed in more detail in subsequent sections. The subtype counterpart to this, Type III-B or Cmr, is differentiated by its use of the cmr5 small subunit. The Cmr complex also has an absence of genes for spacer insertion and processing in the form of Cas1, Cas2, and Cas6. There is evidence that these qualities are covered by modulating the rest of the Cas proteins to fill in this dearth of activity (Hale et al., 2014).
The Type IV CRISPR system, known as Csf1, remains something of an enigma. Having a minimal setup of Cas proteins, it lacks all but the multiprotein effector module. But without spacer proteins or other regulatory controllers, it is not well known how it functions. Possibilities include the complex acting as a mobile unit within the cell that immediately attacks foreign genomes upon contact or that it has a set of already genetically inscribed spacers that act as a type of familial memory for targeting invaders. Further investigation is still required to learn more about this CRISPR assemblage (Makarova et al., 2015).
Class 2 (Type II [Cas9], Type V [Cpf1/Cas12a], Type VI [C2c2/Cas13a])
The Types arranged in Class 2 remain the most well-investigated out of any CRISPR classification, especially when it comes to Cas9. The single unit making up Cas9 is a multidomain protein of exceptional heterogeneity that engenders multifaceted activity including crRNA processing, effector complexing, viral cleavage, and even some spacer insertion. In doing so, it takes the place of the multiprotein amalgamation that characterized Class 1 and allows all of that to be done with a single protein (Chylinski et al., 2014).
This formatting carries over to Type V and the Cpf1 gene, now called Cas12a, which works multi-functionally across domains too. It should be noted that this protein, however, only includes the effector binding module and target cleavage. In comparison to the rest, this system has been found to be highly successful in mammalian cells and its focused homology-directed repair that uses sticky ends over blunt ends increases the efficiency of gene incorporation and expression (Zetsche et al., 2015).
The final category is Type VI that has the C2c2 protein, renamed to Cas13a. The specialized identity of this CRISPR system is that it appears to solely target RNA material, completely lacking any DNA nuclease homology. While some of the other CRISPR mechanisms can sometimes be used against RNA, such as with Type III’s modularity, this is the only known Cas protein to be isolated to RNA targeting (Abadayyeh et al., 2016).
As a sum total, CRISPR and its proteins serve as versatile examples of what bacteria and evolution can create. Each individual component is unique in its function and often there are variants found even between individual species. Research in this field will surely be ongoing for many years to come. For now, in the following sections, the structure, behavior, and efficacy of Cas10 in particular will be elucidated and what new publications on it might reveal for the future of scientific progress.
Overview of CRISPR-Cas10
The CRISPR-Cas 10, Type III, system is an endogenous system used by bacteria to eliminate viral genomic material inside of the cell. It is similar to the CRISPR-Cas9, Type II system, but has several unique characteristics that help it function. In the Type III system, Cas6 acts as a nuclease and cleaves crRNA into intermediate segments (Walker et al., Samai et al., 2015). The CRISPR-Cas 10 complex is able to bind this crRNA and utilize it to target viral RNA (see Figure 2). In order to avoid targeting host genomic material, CRISPR-Cas9 requires short PAM sequences which are uncommon to the host genome (Silas et al., 2017; Kazlauskiene et al., 2017).
Although the mechanism for distinguishing self from nonself is unclear, CRISPR-Cas 10 does not use PAM for host recognition (Bari et al., 2017; Silas et al., 2017). Kazlauskiene et al. (2016) suggests that the Cas10 system is transcription dependent, which limits the likelihood that Cas10 will degrade host sequences. Regardless, this opens the door for biological use of this system, since the target sequence does not need a PAM. In addition, Bari et al. (2017) reported that the Cas10 system was more likely to read through mismatches in the target sequence. This may allow targeting of genes even if point mutations occur. Although this system has potential, there is more research that should be done before using this system with confidence. A complete protocol to purify the CRISPR-Cas 10 system was recently published by Chou-Zheng & Hatoum-Aslan (2017).

Figure 2: Types of CRISPR Systems. a. Type I system, or cascade, cleaves hairpins and uses the sequence, crRNA, Cas3 protein, and PAM sequences to target viral RNA. b. Type II system, or Cas9, uses crRNA as well as tracrRNA from viral RNA in complex with Cas9 to target viral sequences guided by PAMs. c. Type III, or Cas10, uses crRNA cut from Cas6 which associates with the Cas10 complex to base pair with viral DNA and RNA (Marraffini, 2015).
Structure and Subunits of CRISPR-Cas10
The CRISPR-Cas 10, Type III, complex is composed of multiple subunits including Csm2, Csm3, Csm4, and Csm5, as well as the Cas10 subunit (Kazlauskiene et al., 2016; Hatoum-Aslan et al.,2014; Walker et al., 2017). These subunits, as well as other differences, allow CRISPR-Cas 10 to have varied function from the well-known CRISPR Cas9 system. Although little is known about Csm2, it is found associating with Csm5 in the backbone of the CRISPR-Cas 10 structure and deletion of this subunit results in loss of function for the entire complex (Liu, Iavarone, & Doudna, 2017). Likewise, Csm4 associates with Csm3 throughout the complex and the loss of Csm4 results in loss of function for CRISPR-Cas10.
The Csm3 subunit has been studied more since it functions as an RNase which aids in targeting RNA (Kazlauskiene et al., 2016; Hatoum-Aslan et al., 2014). Hatoum-Aslan et al. (2014) reported that Csm3 base pairs with the 6 nucleotides on the 3’ end of the crRNA intermediate. This binding is crucial for the stability of the complex as well as aiding in maturation of crRNA. Without Csm3, the CRISPR-Cas 10 complex is not formed. Further investigation showed that Csm3, as well as Csm5, contains G-rich loops which were the source of protein stability for the complex.
Deletion of Csm5 resulted in CRISPR-Cas 10 associating with intermediate crRNA, but was unable to promote mature crRNA which resulted in a non-functional system (Hatoum-Aslan et al., 2014). In the past, Csm5 has been assumed to catalyze the maturation of crRNA because of its location near the 3’ crRNA end (Walker et al., 2017). However, alanine-scanning mutations in Csm5 did not stop crRNA from maturing which suggests a more supportive or structural role for Csm5. In addition to structure, Csm5 also binds to and recruits PNPase, an exonuclease, to the complex (Walker et al., 2017).
The Cas 10 subunit, also known as Csm1 and Cmr2, is a large subunit with various domains, including an HD domain, two alpha-helix domains, and two Palm domains (Kazlauskiene et al., 2017). The HD domain acts as a nuclease and degrades single stranded RNA (Liu, Iavarone, & Doudna, 2017; Kazlauskiene et al., 2017). The Palm domains have various functions, including crRNA biogenesis, generating Csm6 activator, and acting as a cyclic oligoadenylate synthetase (Niewoehner et al., 2017; Kazlauskiene et al., 2017; Hatoum-Aslan et al., 2014). Additionally, the Palm domain contains sequences similar to a polymerase which suggests a similar role for this region of Cas10 (Hatoum-Aslan et al., 2014; Liu, Iavarone, & Doudna, 2017).
Functions of CRISPR-Cas10
While the overall function of all the CRISPR systems is to eliminate foreign genetic material from the cell, different CRISPR systems are able to target different types of genomic material with different efficiencies. Type III is able to identify and cleave both DNA and RNA.
The CRISPR-Cas 10 system has two different subtypes, A and B, also known as Csm and Cmr (Makarova et al., 2011; Lui, Iavarone, & Doudna, 2017). Type III-A uses the HD-domain of Cas10 to cut foreign DNA, while Class B cleaves RNA (Makarova et al., 2011). Not much is known about Type III-B except that it may work in conjunction with CRISPR Type I-F to target phage RNA which lack PAM sequences (Silas et al., 2017). The similar function between Type III and Type I may be attributed to RAMPs (Repeat Associated Mysterious Proteins). The Cas10 complex, as well as the Cas3 complex from Type I, contains these proteins which participate in RNA binding and cleavage (Rouillon et al., 2013).
Interestingly, although the Type III-A mainly targets DNA, it also contains RAMPs which suggests RNA targeting. Samai et al. (2015) found that Type III-A is able to degrade RNA in addition to DNA at a co-transcriptional level. This allows a single complex to target the parent DNA and the RNA transcripts produced from the DNA at the same time. They further identified that the DNA cleavage was transcript dependent and that the non-template DNA strand was targeted (Samai et al., 2015).
In addition to this, Samai et al. (2015) also found that purified Cas10 was able to cleave double stranded DNA even though it primarily cleaves single stranded DNA (Kazlauskiene et al., 2016). Altogether, the CRISPR-Cas 10 system is diverse enough to target multiple forms of genetic material in double and single stranded forms.
The Applications of CRISPR-Cas10
Genome Editing
CRISPR-Cas 10 has many possible implications in biotechnology and genetics research. The related Type II-A CRISPR system (CRISPR-Cas 9) has been a booming topic in recent studies and has shown much success in gene editing. One study by Yang et al. (2014) used CRISPR-Cas 9 nucleases along with sgRNAs (small guide RNAs) to target and edit the genome of rabbits. Specifically, they used the CRISPR-Cas 9 system to edit nine genes: “apolipoprotein E (APOE), cluster of differentiation 36 (CD36), cystic fibrosis transmembrane conductance regulator, low-density lipoprotein receptor (LDLR), apolipoprotein CIII, scavenger receptor class B, member 1 (SCARB1), leptin, leptin receptor, and ryanodine receptor 2 (RyR2)” (Yang et al., 2014).
They were successful in creating knockout rabbits for each of the nine genes, four of which were new knockout lines for this species (CD36, LDLR, RyR2, and APOE). They created knockouts in these genes by using sgRNAs that were designed specifically to target the above nine genes and then introduce the double stranded break, from which non-homologous end joining, an error prone natural repair mechanism, left the genes non-functional. These researchers also addressed the possible off-target implications of using this system for gene editing. They analyzed exon regions in the rabbit genome and did not find any off-target effects, although mutations in untended genome regions still could have occurred.
They believe this low occurrence of off-target effects may be because of the lower concentration of sgRNA used and the fact they used RNA, which has a shorter half-life, rather than plasmid DNA, an important result in the effort to overcome some of the limitations of this system. Overall, gene editing via the CRISPR-Cas 9 system successfully edited genes and made new knockout rabbits species that will be useful in further research on lipid metabolism, atherosclerosis, and cardiac conditions (Yang et al., 2014).
Bacteriophage Editing
Even more than editing the genomes of several different organisms, now including rabbits, some studies are researching the possibility of editing bacteriophages too. Martel and Moineau (2014) were able to create specific point mutations, deletions, and even insertion of an entire functional methyltransferase gene in the genome of the virulent phage 2972 by swapping a dispensable gene orf33 for the methyltransferase gene. Subsequent phage assays demonstrated that the methyltransferase gene was fully functional (Martel & Moineau, 2014). Being able to edit phage genomes will be useful for further research as the function of much of the phage’s genome, over half, is not well understood (Martel & Moineau, 2014).
Another study by Kiro, Shitrit, and Qimron (2014) also focused on phages but instead used a different system, the type I-E CRISPR-Cas system, in studying the lytic E.coli T7 phages. They also utilized CRISPR-Cas in a different manner than gene editing too, instead using it as a counter-selection technique to select for recombinant plasmids that had a clean deletion of the gene 1.7. The plasmids that still contained gene 1.7 were targeted and cleaved by the Type I-E CRISPR-Cas system whereas the desired plasmids lacking gene 1.7 were not.
This study showed that CRISPR-Cas systems can not only edit genomes, but can be a new means for selection in recombinant plasmid studies more than the current methods of testing antibiotic resistance or with X-gal media. Additionally, this study also demonstrated that other types of CRISPR-Cas systems may prove useful in new research along with CRISPR-Cas 9 although much more research is needed for these different types of CRISPR-Cas systems (Kiro, Shitrit, & Qimron, 2014).
Bacteriophage Editing with CRISPR-Cas 10
One new study investigated phage editing with the CRISPR-Cas 10 system. The native S. epidermidis CRISPR-Cas locus, consisting of four repeats, three spacers, and nine genes for the CRISPR-Cas 10 complex was used to edit the phage genome. However, this locus needed to be inserted next to a spacer matching with the phage of interest. The spacer was provided via a plasmid called pcrispr/spcϕ. Together, this formed the first step of their two-step approach and created what they called the targeting strain.
They tested the targeting strain’s CRISPR-Cas 10 system by introducing the phage and confirming that the spacer indeed gave immunity from the phage to S. epidermidis via targeting and subsequent cleavage of the phage genome with CRISPR-Cas 10. Next, they used the targeting strains to create donor strains which contained an additional homologous sequence on a plasmid that matched one derived from the phage. In the donor strain, this extra homologous sequence was able to provide the needed template for homology-directed repair (see Figure 3).
So even though the phage genome was still targeted and cleaved by CRISPR-Cas 10, the extra homologous sequence on the plasmid allowed homology-directed repair to occur which allowed the phages to “escape further cleavage and complete the infection cycle” (Bari et al., 2017). This study demonstrated that CRISPR-Cas 10 can successfully be used for gene editing like studies with other CRISPR-Cas systems, especially Cas9, have concluded. However, future research will be critical since much is unknown about phage genes and the many possible downstream effects that could come from using this gene editing technique.
Another limitation to widespread application of this technique is that in S. epidermidis the CRISPR-Cas 10 system was a native endogenous system so Bari et al. (2017) also studied S. aureus, a host cell lacking a native CRISPR-Cas system, and instead introduced a plasmid with the S. epidermidis CRISPR-Cas 10 complex into S. aureus. The system still was effective in S. aureus, but editing efficiency was much lower than in the study with the native S. epidermidis CRISPR-Cas 10 complex. Finally, another limitation in this approach is the necessity for the spacer selection to be a region that is actively transcribed (Bari et al., 2017).
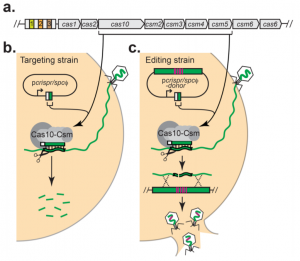
Figure 3: Targeting vs. Editing strain used to edit Staphylococci bacteriophages with CRISPR-Cas 10. a. The native CRISPR-Cas 10 system in S. epidermidis has three repeats (numbered colored rectangles) surrounded by four spacers and several CRISPR-related genes. b. First in the two-step approach was to create a targeting strain using spacer and repeated, provided on the plasmid pcrispr/spcϕ, resulting in immunity from the phage. c. Second, a donor sequence is added to facilitate homology-directed repair and gene editing that allows escape of the Staphylococci bacteriophages from cleavage (Bari et al., 2017).
Conclusion
Overall, it is clear that CRISPR-Cas systems of the several different types can have many important applications in research because of their ability to induce specific gene edits in targeted genes. Even more than simple gene editing, CRISPR-Cas can be used as a new counter-selective method in the creation of recombinant plasmids (Kiro, Shitrit, & Qimron, 2014). Plus, there may be other applications for CRISPR-Cas systems above and beyond gene editing, such as rendering the nuclease domains of the Cas proteins nonfunctional so as to target genes without inducing cleavage. The system could then be used to “carry out new functions such as manipulating gene expression and labeling loci for dynamic cell imaging” (Plummer, Guo, & Peng, 2017).
The possibilities are seemingly endless with the CRISPR-Cas system, but there are several limitations that will need to be attended to. Additionally, research on the specific CRISPR-Cas 10 system and possible applications with gene editing is fairly new and so much more research regarding CRISPR-Cas 10 compared to CRISPR-Cas 9 should be conducted. Since CRISPR-Cas 10 has a more robust immunity against mutant bacteriophages, stemming from the lack of a PAM and seed sequence requirement (Pyenson et al., 2017), CRISPR-Cas 10 could have unique applications compared to other CRISPR-Cas systems that should be further explored.
References
1. Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B. T., … Zhang, F. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science, 353(6299), aaf5573. Retrieved from http://doi.org/10.1126/science.aaf5573
2. Bari, S.M.N., Walker, F.C., Cater, K., Aslan, B., & Hatoum-Aslan, A. (2017). Strategies for editing virulent staphylococcal phages using CRISPR-Cas10. ACS Synthetic Biology. doi:10.1021/acssynbio.7b00240
3. Barrangou, R. (2015). Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biology, 16, 247. Retrieved from http://doi.org/10.1186/s13059-015-0816-9
4. Chou-Zheng, L. & Hatoum-Aslan, A. (2017). Expression and purification of the Cas10-Csm complex from Staphylococci. Bio-Protocol. 7 (11). doi: 10.21769/BioProtoc.2353
5. Chylinski, K., Makarova, K. S., Charpentier, E., & Koonin, E. V. (2014). Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Research, 42(10), 6091–6105. Retrieved from http://doi.org/10.1093/nar/gku241
6. Hale, C. R., Cocozaki, A., Li, H., Terns, R. M., & Terns, M. P. (2014). Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes & Development, 28(21), 2432–2443. Retrieved from http://doi.org/10.1101/gad.250712.114
7. Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., … Terns, M. P. (2009). RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell, 139(5), 945–956. Retrieved from http://doi.org/10.1016/j.cell.2009.07.040
8. Hatoum-Aslan, A., Maniv, I., Samai, P., & Marraffini, L. A. (2014). Genetic Characterization of Antiplasmid Immunity through a Type III-A CRISPR-Cas System. Journal of Bacteriology, 196 (2), 310-317. doi:10.1128/JB.01130-13
9. Jinek, M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337(6096), 816–821. Retrieved from http://doi.org/10.1126/science.1225829
10. Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., … Brouns S.J. (2011). Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Structural & Molecular Biology, 18, 529–536. Retrieved from http://doi.org/10.1038/nsmb.2019
11. Kazlauskiene, M., Kostiuk, G., Venclovas, Č., Tamulaitis, G., & Siksnys, V. (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 11, 357 (6351), 605-609. doi: 10.1126/science.aao0100
12. Kazlauskiene, M., Tamulaitis, G., Kostiuk, G., Venclovas, Č., & Siksnys, V. (2016). Spatiotemporal control of type III-A CRISPR-Cas immunity: Coupling DNA degradation with the target RNA recognition. Molecular Cell, 62 (2), 295-306. doi: 10.1016/j.molcel.2016.03.024
13. Kiro, R., Shitrit, D., & Qimron, U. (2014). Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biology, 11(1), 42–44. Retrieved from http://doi.org/10.4161/rna.27766
14. Liu, T.Y., Iavarone, A.T., Doudna, J.A. (2017). RNA and DNA Targeting by a reconstituted thermus thermophilus type III-A CRISPR-Cas system. PLoS One, 12 (1). doi: 10.1371/journal.pone.0170552
15. Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., … Koonin, E. V. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nature Reviews. Microbiology, 13(11), 722–736. Retrieved from http://doi.org/10.1038/nrmicro3569
16. Makarova, K.S., Haft, D.H., Barrangou, R., Brouns, S.J., Charpentier, E., Horvath, P… Koonin, E.V. (2011). Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol, 9(6):467–77. doi: 10.1038/nrmicro2577
17. Marraffini, L. A. (2015). CRISPR-Cas Systems in Prokaryotes [Image]. Nature, 526, 55-61. doi:10.1038/nature15386
18. Martel, B., & Moineau, S. (2014). CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Research, 42(14), 9504–9513. Retrieved from http://doi.org/10.1093/nar/gku628
19. Niewoehner, O., Garcia-Doval, C., Rostøl, J.T., Berk, C., Schwede, F., Bigler, L., Hall, J., Marraffini, L.A., & Jinek, M. (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature, 548 (7669), 543-548. doi: 10.1038/nature23467
20. Plummer, R.J., Guo, Y., & Peng, Y. (2017). A CRISPR reimagining: new twists and turns of CRISPR beyond the genome-engineering revolution. J Cell Biochem. Doi: 10.1002/jcb.26406
21. Pyenson, N.C., Gayvert, K., Varble, A., Elemento, O., & Marraffini, L. A. (2017). Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host and Microbe, 22 (3), 343-353. Retrieved from http://dx.doi.org/10.1016/j.chom.2017.07.016
22. Rouillon, C., Zhou, M., Zhang, J., Politis, A., Beilsten-Edmands, V., Cannone, G., … White, M. F. (2013). Structure of the CRISPR Interference Complex CSM Reveals Key Similarities with Cascade. Molecular Cell, 52(1), 124–134. http://doi.org/10.1016/j.molcel.2013.08.020
23. Samai, P., Pyenson, N., Jiang, W., Goldberg, G. W., Hatoum-Aslan, A., & Marraffini, L. A. (2015). Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell, 161, 1164-1174. doi:10.1016/j.cell.2015.04.027
24. Sashital, D. G., Wiedenheft, B., & Doudna, J. A. (2012). Mechanism of foreign DNA selection in a bacterial adaptive immune system. Molecular Cell, 46(5), 606–615. Retrieved from http://doi.org/10.1016/j.molcel.2012.03.020
25. Silas, S., Lucas-Elio, P., Jackson, S. A., Aroca-Crevillén, A., Hansen, L. L., Fineran, P. C., … Sánchez-Amat, A. (2017). Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. eLife, 6, e27601. http://doi.org/10.7554/eLife.27601
26. Walker, F.C., Chou-Zheng, L., Dunkle, J.A., Hatoum-Aslan, A. (2017). Molecular determinants for CRISPR RNA maturation in the Cas10-Csm complex and roles for non-Cas nucleases. Nucleic Acids Research, 45 (4), 2112-2123. doi: 10.1093/nar/gkw891
27. Wiedenheft, B., Lander, G. C., Zhou, K., Jore, M. M., Brouns, S. J. J., van der Oost, J., … Nogales, E. (2011). Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature, 477(7365), 486–489. Retrieved from http://doi.org/10.1038/nature10402
28. Yang, D., Xu, J., Zhu, T., Fan, J., Lai, L., Zhang, J., & Chen, Y. E. (2014). Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. Journal of Molecular Cell Biology, 6(1), 97–99. http://doi.org/10.1093/jmcb/mjt047
29. Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., … Zhang, F. (2015). Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell, 163(3), 759–771. Retrieved from http://doi.org/10.1016/j.cell.2015.09.038
Photo CCs: Phi X 174 slice from Wikimedia Commons





